Blog | Reading Time 2 minutes
How to study the microbiome?
In recent years, the advances and increased affordability of DNA sequencing techniques have allowed scientists to grow our understanding of the animal microbiome, but how does this work?
What is metagenomics?
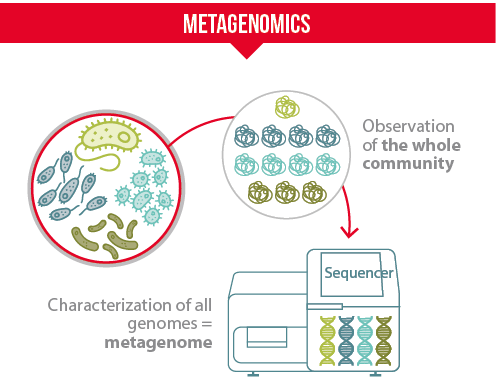
Microbiologists used to apply cultural techniques and microscopy to identify and characterize microorganisms. These techniques are time-consuming and provide a limited vision of a microbial community. Lately, thanks to the progress of high-throughput sequencing technologies, microbiologists have made a giant leap with the development of metagenomics.
- Metagenomics applies a suite of sequencing technologies and bioinformatics tools to directly access the genetic content of entire communities of microorganisms (Thomas et al., 2012). Cultural and metagenomics approaches are complementary.
What characterize metagenomics techniques:
- They allow detecting rare bacteria species and ones that cannot be cultivated
- They enable the analysis of a large number of microbial samples at the same time
- They give a snapshot of populations diversity within a sample
These techniques require very specific expertise in biostatistics and important bioinformatics to translate the millions of DNA sequences generated into microbial population composition. They also rely on a specific database of known bacteria sequences, while a large proportion is still unknown. Depending on the level of detail needed and the complexity of the experimental design, it can take between one week to several months to analyze a sequencing dataset.
From metagenomics to barcoding: 16S rRNA
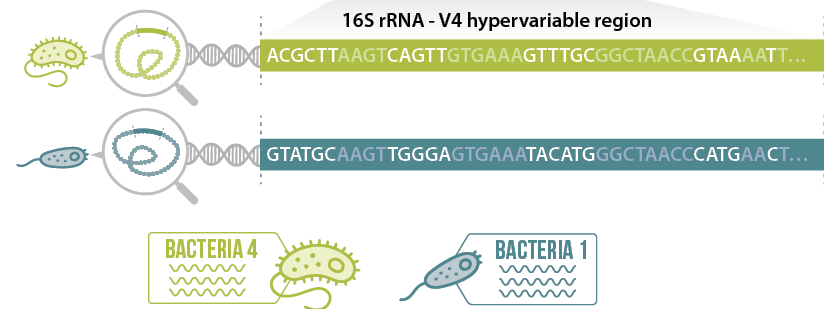
To describe the microbial composition of gut microbiota using sequencing strategy it is not necessary to sequence the full metagenome. A common practice is to target a fragment of the bacterial genome, which is used as a marker or a sort of ID card of a bacteria. This approach, called amplicon sequencing or barcoding, drastically reduces the cost and the time of analysis
- The 16S ribosomal RNA subunit gene (16S rRNA gene) is the most used marker gene to describe bacteria composition.
The rapid and substantial cost reduction in next-generation sequencing has dramatically accelerated the development of metagenomics and the understanding of host-microbiota.
Published Aug 13, 2021 | Updated May 30, 2023
Related articles
Need specific information?
Talk to an expert